According to Nature, researchers have identified eight promising drug candidates targeting the SARS-CoV-2 NSP6 protein using advanced computational methods including structure prediction, molecular docking, and 100-nanosecond molecular dynamics simulations. The study screened over 2.9 million compounds from the ZINC20 database and identified candidates with favorable drug-like properties and binding stability to NSP6’s critical regions. This computational approach represents a significant step toward developing next-generation COVID-19 treatments.
Table of Contents
Understanding NSP6’s Role in Viral Replication
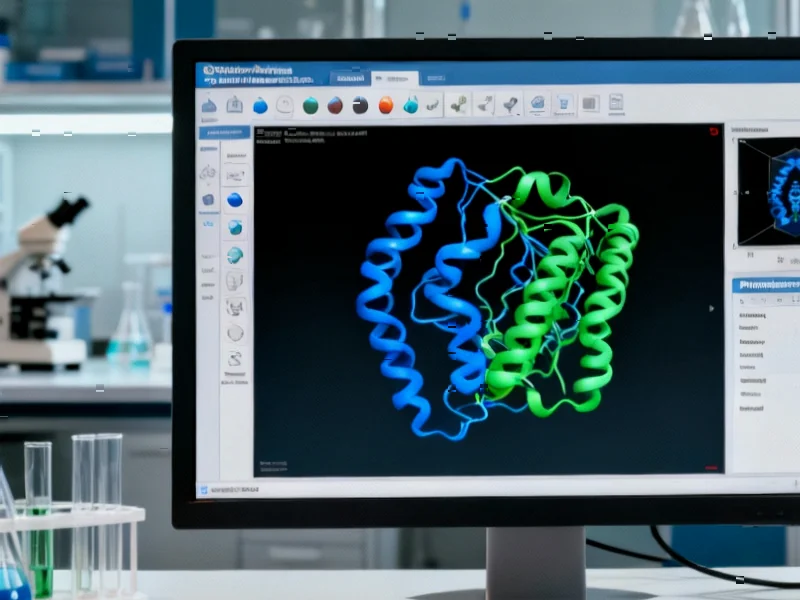
NSP6 is one of the non-structural proteins encoded by the SARS-CoV-2 genome that plays a crucial role in creating double-membrane vesicles where viral replication occurs. Unlike the spike protein that has been the primary target for vaccines and antibodies, NSP6 operates behind the scenes in viral replication machinery. The protein’s structure, characterized by eight transmembrane helices and significant alpha-helical content, makes it challenging to target with conventional small molecules. The researchers’ use of protein secondary structure prediction tools revealed NSP6 is composed of 77.93% α-helices, which are stable structural elements that can form effective binding pockets for potential drugs.
Critical Analysis of Computational Drug Discovery
While the computational approach is impressive in scale and sophistication, several critical challenges remain before these findings can translate to clinical treatments. The 100-nanosecond simulation, while substantial for computational studies, represents only a fraction of the time scales relevant to biological systems. More importantly, NSP6 functions within complex cellular environments and interacts with other viral proteins and host factors that weren’t accounted for in these isolated simulations. The study’s focus on molecular docking and binding stability doesn’t address whether disrupting NSP6 function alone would sufficiently inhibit viral replication, given the virus’s redundant mechanisms and adaptive capabilities.
Validation Challenges and Next Steps
The transition from computational hits to viable drugs faces substantial hurdles that the study doesn’t address. Laboratory validation using cellular assays and animal models is essential to confirm that these compounds actually inhibit viral replication without significant toxicity. The compounds’ ability to penetrate cells and reach intracellular compartments where NSP6 functions remains unverified. Additionally, the study doesn’t consider potential off-target effects or interactions with human proteins, which could lead to unexpected side effects. The promising binding energies and stability metrics need correlation with actual antiviral activity before these candidates can progress to preclinical development.
Broader Implications for Antiviral Development
This research demonstrates the growing power of computational methods in pandemic preparedness and rapid drug discovery. Targeting non-structural proteins like NSP6 represents a strategic shift from the spike-protein-focused approaches that dominated early COVID-19 research. If successful, such compounds could work against multiple coronavirus variants since NSP6 is more conserved across variants than the frequently mutating spike protein. The methodology also showcases how alpha helix-rich transmembrane proteins, traditionally difficult drug targets, are becoming more accessible through advanced computational techniques. This could open doors for targeting similar proteins in other viral diseases.
Realistic Outlook and Future Directions
The identification of these eight candidates represents the very beginning of a long drug development pathway that typically takes years and has high failure rates. However, the systematic approach combining multiple computational validation methods increases confidence in these initial hits. The most promising aspect is that several candidates showed stable binding to regions containing predicted T-cell epitopes, suggesting they might interfere with immune recognition as well as structural function. As computational power increases and algorithms improve, we can expect more sophisticated simulations that better replicate cellular conditions and accelerate the transition from virtual screening to viable clinical candidates for future pandemic threats.